|
Customising Annotations | ||||||
In this tutorial we will demonstrate how you can display all the Genbank identifiers for the Human (NlaIII) SAGE tags. This is done by defining how the SAGE tags are 'related' to the genbank entries. The relationship is defined by a series of mappings between the different identifiers, in this case we state that to get the GI entries for SAGE tags we:
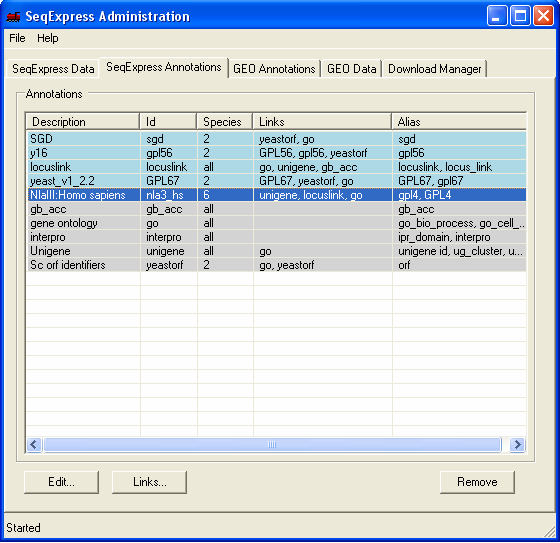
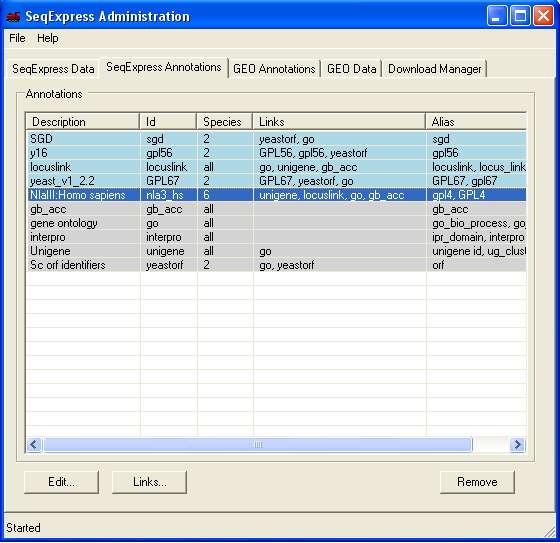
The SeqExpress Annotations tab shows information about all currently loaded annotations. Presently the SAGE tags have links to unigene, locuslink and GO (so for each SAGE tag we can lookup any of these corresponding annotations). To add a the genbank mapping, select the SAGE tag row, and click the Links button.
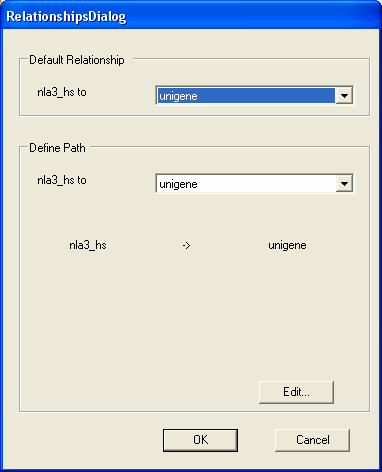
A dialog appears showing at the top the name of the default annotations that are to be displayed for SAGE tags (in this case the unigene entries). Below that is information about how the unigene entries for the SAGE tags are worked out (in this case information about unigene to SAGE mappings are directly available);
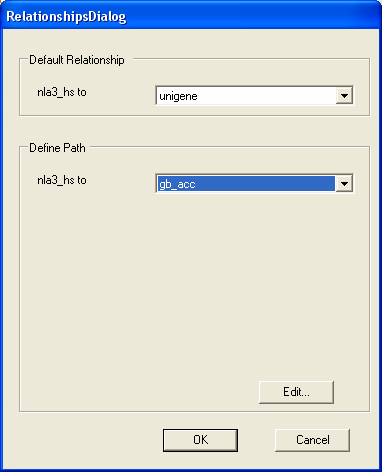
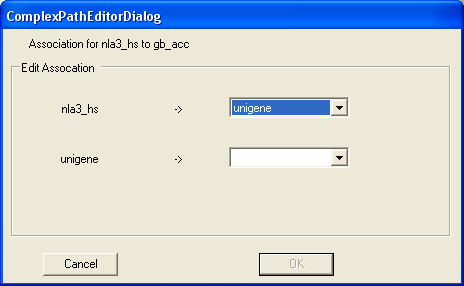
To define the SAGE to Genbank mapping you should first select the gb_acc annotation from the define path box. Then select the Edit button (as we are now going to edit how the genbank identifiers are looked up for specific SAGE tags).
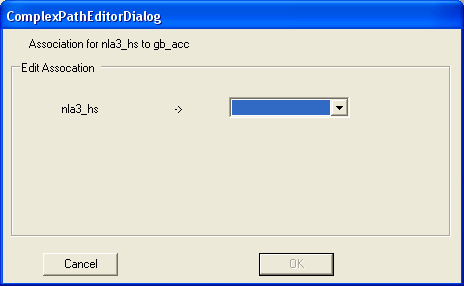
The relationships between SAGE and Genbank is defined by using annotation mappings that have already been loaded into SeqExpress.
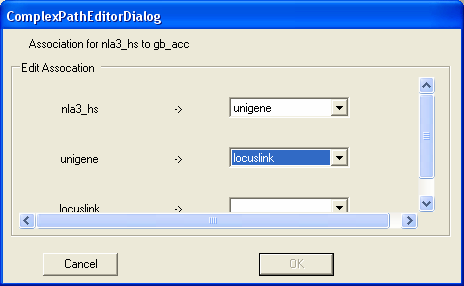
First we wish to navigate from SAGE tags to unigene clusters.
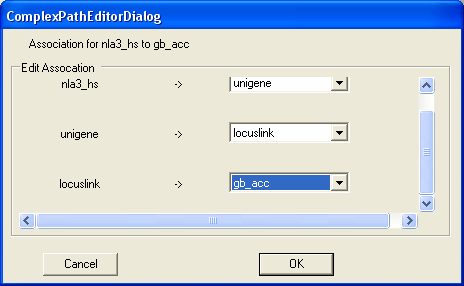
Then we wish to navigate from unigene clusters to locuslink entries
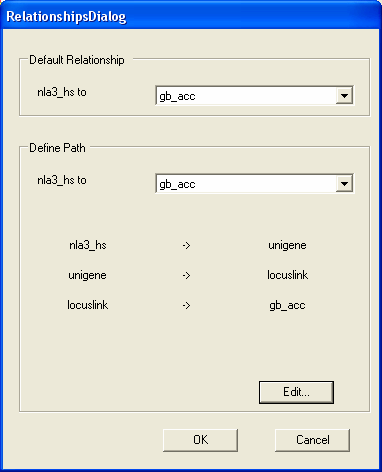
Then we wish to navigate from locuslink entries to genbank identifiers. As the relationship is now defined you should select the OK button.
The relationship between SAGE tags and GI entries is now established. It is also now the default relationship, so that when Human NlaIII SAGE tags are loaded into SeqExpress the corresponding genbank identifiers will be shown. You can change the annotations that are shown in SeqExpress at runtime by using the Edit->Annotations menu option. You should select OK to continue.
The new navigation has been defined, and we can see a new link between the nla3_hs annotation type and gb_acc.
|